Inhibition of SARS-CoV-2-mediated thromboinflammation by CLEC2.Fc
- Details
- Published: Monday, 05 June 2023 12:00
With the rampant spread of the novel coronavirus (SARS-CoV-2) worldwide, it has caused severe inflammation and even death in COVID-19 patients. Even after recovery, there is still a possibility of contracting long COVID, which has long-lasting and serious impacts on the lives of diagnosed individuals. Thromboinflammation refers to the combination of blood clotting and inflammation, and it is the primary cause of disease progression and mortality in COVID-19 patients. Autopsies have shown the presence of platelet-rich clots and microangiopathy (abnormalities in small blood vessels) in various organs. Furthermore, studies have found persistent microclots in the plasma of both acute COVID-19 and long COVID patients. However, the exact molecular mechanism of SARS-CoV-2-induced thromboinflammation remains unclear.
Dr. Shie-Liang Edmond Hsieh and his research team have discovered a direct interaction between spleen tyrosine kinase (Syk)-coupled receptor CLEC2, which is highly expressed in platelets and alveolar macrophages, and the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein. They have observed that SARS-CoV-2 induces a different type of neutrophil extracellular trap (NET) formation, known as aggregated NETs, in the presence of platelets. Moreover, by stimulating platelets and neutrophils with lentivirus expressing the RBD of the SARS-CoV-2 spike protein, they have confirmed that SARS-CoV-2 activates platelets through the binding of RBD and CLEC2, leading to increased NET formation and thromboinflammation(Figure1).
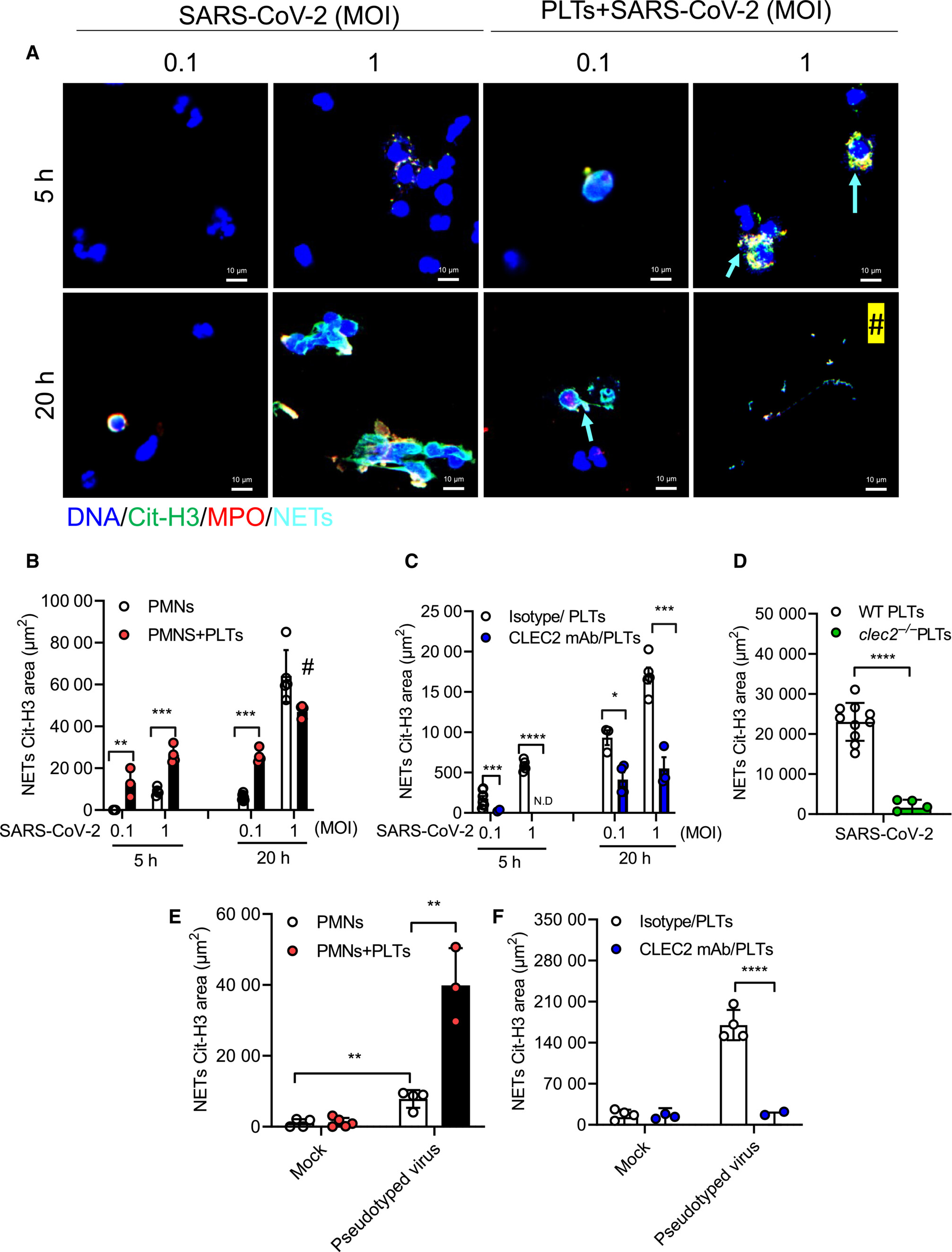 |
| Figure 1: SARS-CoV-2 activates platelet CLEC2 to induce NET formation via spike RBD |
To evaluate the potential therapeutic implications of these findings, the research team administered recombinant CLEC2 protein (CLEC2.Fc) to mice infected with SARS-CoV-2 (AAV-hACE2-infected mice). They found that recombinant CLEC2.Fc effectively inhibited the formation of NETs in the lungs and hearts induced by SARS-CoV-2 infection, and reduced thromboinflammation in the mice (Figure 2).
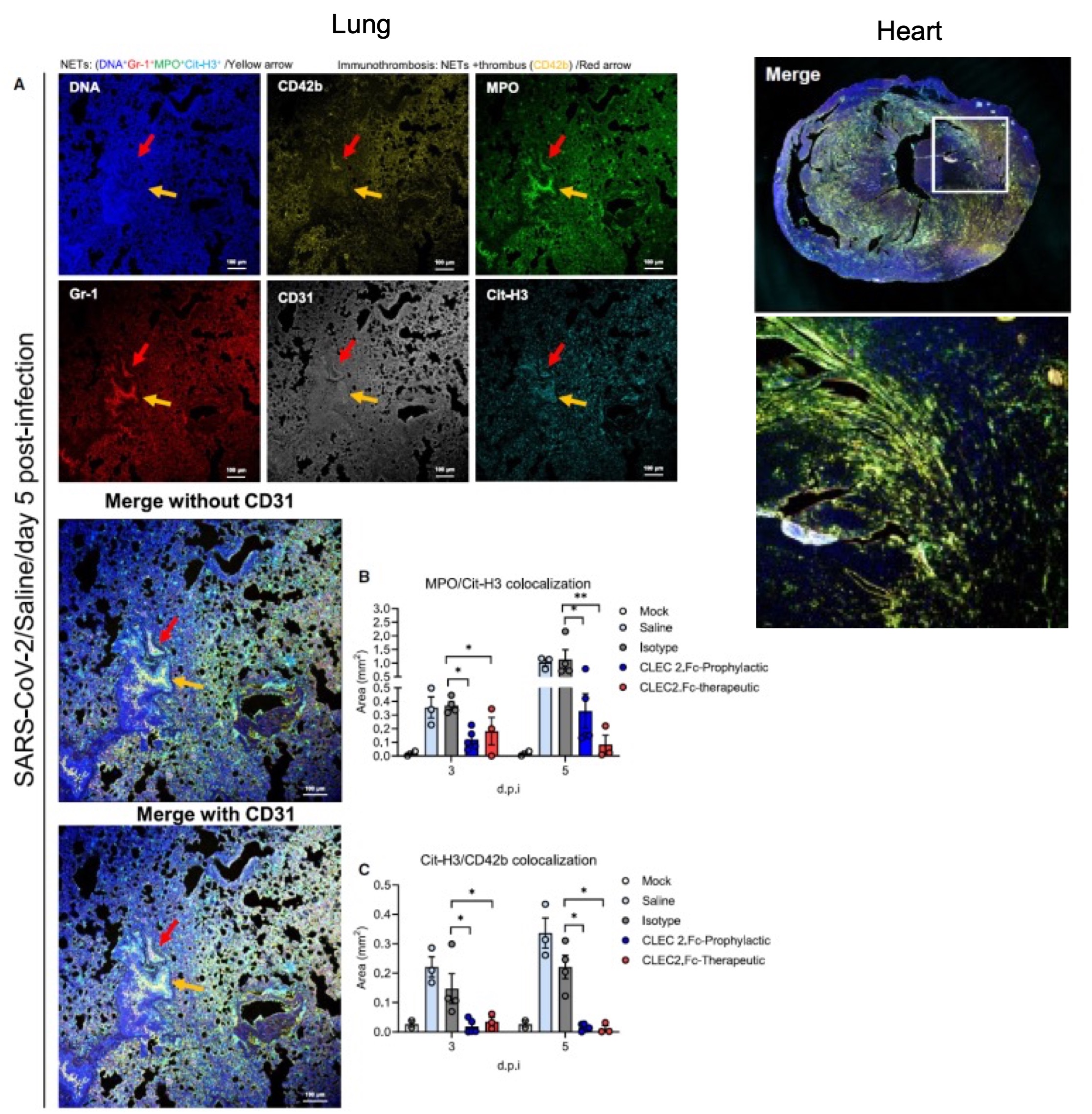 |
| Figure 2. CLEC2.Fc inhibits SARS-CoV-2-induced NETs and thrombosis in the lungs and hearts in SARS-CoV-2-infected mice. |
This suggests that CLEC2 serves as a novel pattern recognition receptor for SARS-CoV-2, and CLEC2.Fc is a promising therapeutic agent to counteract SARS-CoV-2-induced thromboinflammation and potentially reduce the risk of post-acute sequelae of COVID-19 (PASC). The molecular mechanism was shown in Figure 3.
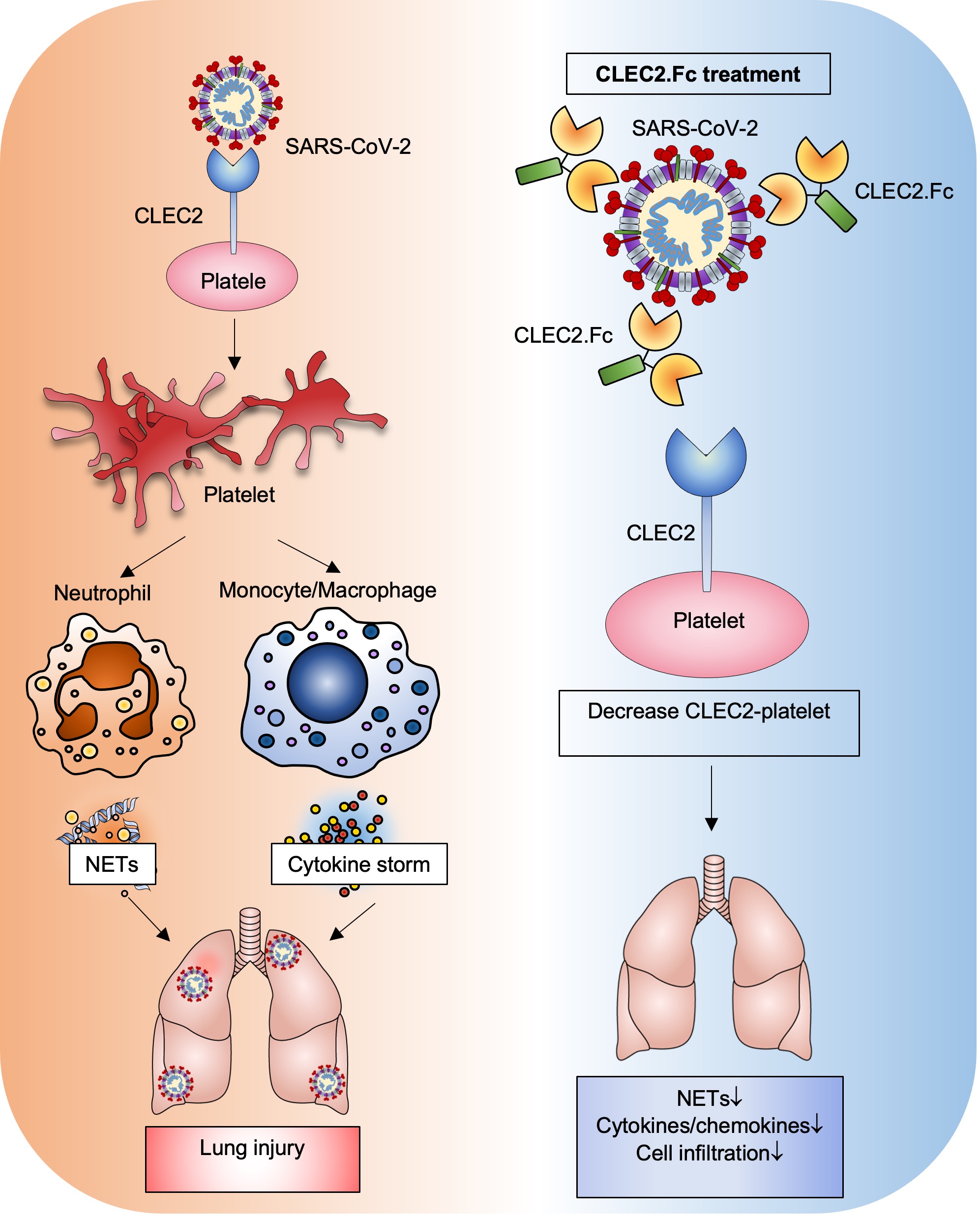 |
| Figure 3: CLEC2.Fc-mediated anti-inflammatory effects in vivo may be via blocking: a) SARS-CoV-2-induced platelet activation and cytokine secretion from activated macrophages; and b) podoplanin-mediated inflammatory reactions in vivo. |
A full study “Inhibition of SARS-CoV-2-mediated thromboinflammation by CLEC2.Fc” is published recently in the EMBO Molecular Medicine, in which Dr. Pei-Shan Sung, a post-doctoral fellow at GRC, is the first author. This article can be read online at: https://www.embopress.org/doi/full/10.15252/emmm.202216351