 Professor
Professor
yangas
+886-2-27871232
+886-2-27871232
EDUCATION AND POSITIONS HELD:
- Ph.D., Department of Chemistry The Johns Hopkins University, 1987
- Postdoctoral Fellow, Department of Physics University of Virginia, 1987–1988
- Postdoctoral Fellow, Department of Biochemistry Columbia University, 1988–1991
- Associate Research Scientist - Research Scientist, Columbia University, 1991–2000
- Assistant Professor of Pharmacology, Columbia University, 2000–2004
- Associate Professor, Genomics Research Center, Academia Sinica, 2004 - 2010
- Deputy Director, Genomics Research Center, Academia Sinica, 2007 - 2016
- Professor, Genomics Research Center, Academia Sinica, 2010 - 2023
- Division director of the Physical and Computational Genomics division, Genomics Research Center, Academia Sinica, 2018 – 2021
HONORS:
- Fellowship (2000-2002), William J. Matheson foundation at Columbia University.
RESEARCH INTERESTS:
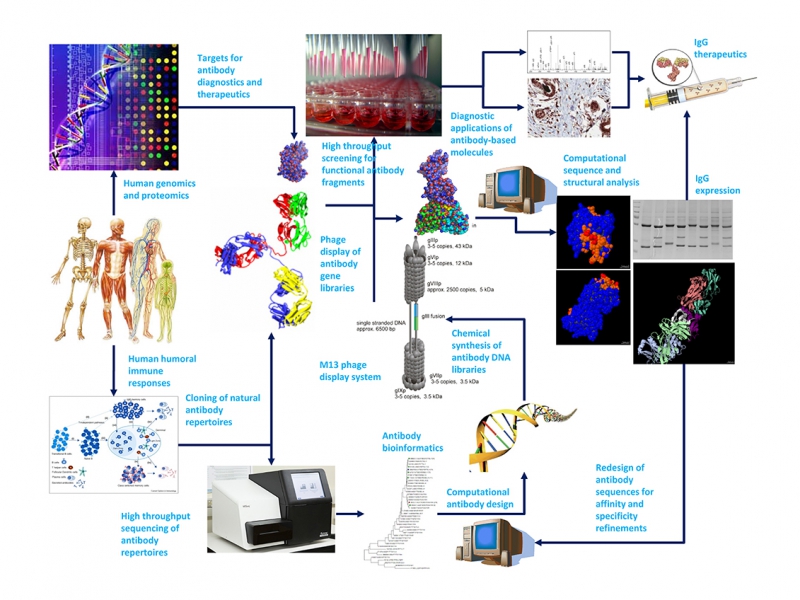 Our lab has implemented phage-based protein display systems in connection with computational and bioinformatics methodologies harnessing the power of rapidly expanding computing capabilities and high throughput experimental technologies for antibody/protein engineering and antibody/protein design. The research directions are aiming at innovating antibody/protein design and engineering technologies and understanding the biological function of natural antibody repertoires with synthetic phage-displayed antibody libraries and bioinformatics; the goal is to develop antibody-based molecules for important biomedical applications. The works from our group can be found in [https://scholar.google.com.tw/citations?user=YBoSfnAAAAAJ&hl=en]
Our lab has implemented phage-based protein display systems in connection with computational and bioinformatics methodologies harnessing the power of rapidly expanding computing capabilities and high throughput experimental technologies for antibody/protein engineering and antibody/protein design. The research directions are aiming at innovating antibody/protein design and engineering technologies and understanding the biological function of natural antibody repertoires with synthetic phage-displayed antibody libraries and bioinformatics; the goal is to develop antibody-based molecules for important biomedical applications. The works from our group can be found in [https://scholar.google.com.tw/citations?user=YBoSfnAAAAAJ&hl=en]
SELECTED PUBLICATIONS:
- Hung-Pin Peng, An-Suei Yang*, accepted, “Computational Analysis of Antibody Paratopes for Antibody Sequences in Antibody Libraries”, editor(s): Daisuke Kuroda and Kouhei Tsumoto (Eds), Methods Molecular Biology, Vol. 2552, : Computer-Aided Antibody Design, pp. Chapter 24, Springer Nature.
- Peng Hung-Pin, Hsu Hung-Ju, Yu Chung-Ming, Hung Fei-Hung, Tung Chao-Ping, Huang Yu-Chuan, Chen Chi-Yung, Tsai Pei-Hsun, Yang An-Suei*, 2022, “Antibody CDR amino acids underlying the functionality of antibody repertoires in recognizing diverse protein antigens”, Scientific Reports, 12(1), 12555. (SCIE)
- Forinová Michala, Pilipenco Alina, Víšová Ivana, Lynn N. Scott, Dostálek Jakub, Mašková Hana, Hönig Václav, Palus Martin, Selinger Martin, Kočová Pavlína, Dyčka Filip, Štěrba Jan, Houska Milan, Vrabcová Markéta, Horák Petr, Anthi Judita, Tung Chao-Ping, Yu Chung-Ming, Chen Chi-Yung, Huang Yu-Chuan, Tsai Pei-Hsun, Lin Szu-Yu, Hsu Hung-Ju, Yang An-Suei, Dejneka Alexandr, Vaisocherová-Lísalová Hana*, 2021, “Functionalized Terpolymer-Brush-Based Biointerface with Improved Antifouling Properties for Ultra-Sensitive Direct Detection of Virus in Crude Clinical Samples”, ACS Applied Materials & Interfaces, 13(50), 60612-60624. (SCIE)
- Ko YA, Yu YH, Wu YF, Tseng YC, Chen CL, Goh KS, Liao HY, Chen TH, Cheng TR, Yang AS, Wong CH, Ma C, Lin KI, 2021, “A non-neutralizing antibody broadly protects against influenza virus infection by engaging effector cells.”, PLoS pathogens, 17(8), e1009724. (SCIE)
- Hsu HJ, Tung CP, Yu CM, Chen CY, Chen HS, Huang YC, Tsai PH, Lin SI, Peng HP, Chiu YK, Tsou YL, Kuo WY, Jian JW, Hung FH, Hsieh CY, Hsiao M, Chuang SS, Shen CN, Wang YA, Yang AS*, 2021, “Eradicating mesothelin-positive human gastric and pancreatic tumors in xenograft models with optimized anti-mesothelin antibody-drug conjugates from synthetic antibody libraries.”, Scientific reports, 11(1), 15430. (SCIE)
- Tsai Tsung-Yu, Huang Ming-Ting, Sung Pei-Shan, Peng Cheng-Yuan, Tao Mi-Hua, Yang Hwai-I, Chang Wei-Chiao, Yang An-Suei, Yu Chung-Ming, Lin Ya-Ping, Bau Ching-Yu, Huang Chih-Jen, Pan Mei-Hung, Wu Chung-Yi, Hsiao Chwan-Deng, Yeh Yi-Hung, Duan Shiteng, Paulson James C, Hsieh Shie-Liang, 2021, “SIGLEC-3 (CD33) serves as an immune checkpoint receptor for HBV infection”, Journal of Clinical Investigation, 131(11), e141965. (SCIE)
- Huang YL, Huang MT, Sung PS, Chou TY, Yang RB, Yang AS, Yu CM, Hsu YW, Chang WC, Hsieh SL, 2021, “Endosomal TLR3 co-receptor CLEC18A enhances host immune response to viral infection.”, Communications biology, 4(1), 229. (SCIE)
- Kuo WC, Lee CC, Chang YW, Pang W, Chen HS, Hou SC, Lo SY, Yang AS, Wang AH*, 2021, “Structure-based Development of Human Interleukin-1beta-Specific Antibody That Simultaneously Inhibits Binding to Both IL-1RI and IL-1RAcP.”, Journal of molecular biology, 433(4), 166766. (SCIE)
- Chiang ZC, Chiu YK, Lee CC, Hsu NS, Tsou YL, Chen HS, Hsu HR, Yang TJ, Yang AS, Wang AH*, 2020, “Preparation and characterization of antibody-drug conjugates acting on HER2-positive cancer cells.”, PloS one, 15(9), e0239813. (SCIE)
- Chung-Ming Yu, Ing-Chien Chen, Chao-Ping Tung, Hung-Pin Peng, Jhih-Wei Jian, Yi-Kai Chiu, Yueh-Liang Tsou, Hong-Sen Chen, Yi-Jen Huang, Wesley Wei-Wen Hsiao, Yong Alison Wang, An-Suei Yang*, 2020, “A panel of anti-influenza virus nucleoprotein antibodies selected from phage-displayed synthetic antibody libraries with rapid diagnostic capability to distinguish diverse influenza virus subtypes”, SCIENTIFIC REPORTS, 10(1), 13318. (SCIE)
- Fei-Hung Hung, Yong Alison Wang*, Jhih-Wei Jian , Hung-Pin Peng , Ling-Ling Hsieh , Chen-Fang Hung , Max Yang , An-Suei Yang*, 2019, “Evaluating BRCA mutation risk predictive models in a Chinese cohort in Taiwan”, SCIENTIFIC REPORTS, 9(1), 10229. (SCIE)
- Jhih-Wei Jian, Hong-Sen Chen, Yi-Kai Chiu, Hung-Pin Peng, Chao-Ping Tung, Ing-Chien Chen, Chung-Ming Yu, Yueh-Liang Tsou, Wei-Ying Kuo, Hung-Ju Hsu, An-Suei Yang*, 2019, “Effective binding to protein antigens by antibodies from antibody libraries designed with enhanced protein recognition propensities”, MABS, 11(2): 373-387. (SCIE)
- Wei-Ying Kuo, Hung-Ju Hsu, Chun-Yi Wu, Hong-Sen Chen, Yu-Chi Chou, Yueh-Liang Tsou, Hung-Pin Peng, Jhih-Wei Jian, Chung-Ming Yu, Yi-Kai Chiu, Ing-Chien Chen, Chao-Ping Tung, Michael Hsiao, Chia-Lung Lin, Yong Alison Wang, Andrew H-J. Wang, An-Suei Yang*, 2018, “Antibody-drug conjugates with HER2-targeting antibodies from synthetic antibody libraries are highly potent against HER2 positive human gastric tumor in xenograft models”, mAbs, 11(1), 153-165. (SCIE)
- Wei-Ying Kuo, Jia-Jia Lin, Hung-Ju Hsu, Hong-Sen Chen, An-Suei Yang*, and Chun-Yi Wu*, 2018, “Noninvasive assessment of characteristics of novel anti-HER2 antibodies by molecular imaging in a human gastric cancer xenograft-bearing mouse model”, SCIENTIFIC REPORTS, 8(1), 13735. (SCIE)
- Yong Alison Wang*, Jhih-Wei Jian, Chen-Fang Hung, Hung-Pin Peng, Chi-Fan Yang, Hung-Chun Skye Cheng, An-Suei Yang*, 2018, “Germline breast cancer susceptibility gene mutations and breast cancer outcomes”, BMC CANCER, 18(1), 315-327. (SCIE)
- Ing-Chien Chen, Yi-Kai Chiu, Chung-Ming Yu, Cheng-Chung Lee, Chao-Ping Tung, Yueh-Liang Tsou, Yi-Jen Huang, Chia-Lung Lin, Hong-Sen Chen, Andrew H.-J. Wang, An-Suei Yang*, 2017, “High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries”, SCIENTIFIC REPORTS, 7(1), 14455. (SCIE)
- Kwong PD, Chuang GY, DeKosky BJ, Gindin T, Georgiev IS, Lemmin T, Schramm CA, Sheng Z, Soto C, Yang AS, Mascola JR, Shapiro L, 2017, “Antibodyomics: bioinformatics technologies for understanding B-cell immunity to HIV-1.”, Immunological reviews, 275(1), 108-128. (SCIE)
- Hou SC, Chen HS, Lin HW, Chao WT, Chen YS, Fu CY, Yu CM, Huang KF, Wang AH, Yang AS*, 2016, “High throughput cytotoxicity screening of anti-HER2 immunotoxins conjugated with antibody fragments from phage-displayed synthetic antibody libraries.”, Scientific reports, 6, 31878. (SCIE)
- Jian JW, Elumalai P, Pitti T, Wu CY, Tsai KC, Chang JY, Peng HP, Yang AS*, 2016, “Predicting Ligand Binding Sites on Protein Surfaces by 3-Dimensional Probability Density Distributions of Interacting Atoms.”, PloS one, 11(8), e0160315. (SCIE)
- Chao-Ping Tung, Ing-Chien Chen, Chung-Ming Yu, Hung-Pin Peng, Jhih-Wei Jian, Shiou-Hwa Ma, Yu-Ching Lee, Jia-Tsrong Jan, An-Suei Yang*, 2015, “Discovering neutralizing antibodies targeting at the stem epitope of influenza hemagglutinin with synthetic phage-displayed antibody libraries”, Scientific Reports, 5, 15053. (SCIE)
- Chen, H.-S., S.-C. Hou, J.-W. Jian, K.-S. Goh, S.-T. Shen, Y.-C. Lee, J.-J. You, H.-P. Peng, W.-C. Kuo, S.-T. Chen, M.-C. Peng, H.-J. Wang, C.-M. Yu, I.-C. Chen, C.-P. Tung, T.-H. Chen, K. P. Chiu, C. Ma, C. Y. Wu, S.-W. Lin and A.-S. Yang*, 2015, “Predominant structural configuration in natural antibody repertoires enables potent antibody responses against protein antigens”, Scientific Reports, 5, 12411. (SCIE)
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O'Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD, 2015, “Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.”, Cell, 161, 1280-1292. (SCIE)
- Hung-Pin Peng, Kuo Hao Lee, Jhih-Wei Jian, An-Suei Yang*, 2014, “Origins of specificity and affinity in antibody-protein interactions”, PROCEEDINGS OF THE NATIONAL ACADEMY OF SCIENCES OF THE UNITED STATES OF AMERICA, 111(26), E2656-E2665. (SCIE)
- Rajasekaran Mahalingama*, Hung-Pin Peng, An-Suei Yang*, 2014, “Prediction of fatty acid-binding residues on protein surfaces with three-dimensional probability distributions of interacting atoms”, BIOPHYSICAL CHEMISTRY, 192, 10-19. (SCIE)
- Rajasekaran Mahalingam*, Hung-Pin Peng, An-Suei Yang*, 2014, “Prediction of FMN-binding residues with three-dimensional probability distributions of interacting atoms on protein surfaces”, JOURNAL OF THEORETICAL BIOLOGY, 343, 154-161. (SCIE)
- Hsu, H.-J., K. H. Lee, J.-W. Jian, H.-J. Chang, C.-M. Yu, Y.-C. Lee, I.-C. Chen, H.-P. Peng, C. Y. Wu, Y.-F. Huang, C.-Y. Shao, K. P. Chiu and A.-S. Yang*, 2014, “Antibody variable domain interface and framework sequence requirements for stability and function by high throughput experiments.”, STRUCTURE, 22(1), 22-34. (SCIE)
- Chang, H.-J., J.-W. Jian, H.-J. Hsu, Y.-C. Lee, H.-S. Chen, J.-J. You, S.-C. Hou, C.-Y. Shao, Y.-J. Chen, K.-P. Chiu, H.-P. Peng, K. H. Lee and A.-S. Yang*, 2014, “Loop sequence features and stability determinants in antibody variable domains by high throughput experiments.”, STRUCTURE, 22(1), 9-21. (SCIE)
- Chang, H.-J. and A.-S. Yang*, 2014, “Design of Phage-Displayed Cystine-Stabilized Mini-Protein Libraries for Proteinaceous Binder Engineering”, Methods in Molecular Biology, 1088, 1-17.
- Hsu, H.-J. and A.-S. Yang*, 2014, “Substrate Phage Display for Protease Substrate Sequence Characterization: Bovine Factor Xa as a Model System”, Methods in Molecular Biology, 1088, 107-124.
- Keng-Chang Tsai, Ei-Wen Yang, Jhih-Wei Jian, Po-Chiang Hsu, Hung-Pin Peng, Ching-Tai Chen, Jun-Bo Chen, Jeng-Yih Chang, Wen-Lian Hsu. An-Suei Yang*, 2012, “Prediction of Carbohydrate Binding Sites on Protein Surfaces with 3-Dimensional Probability Density Distributions of Interacting Atoms”, PLoS One, 7(7), e40846. (SCIE)
- Ching-Tai Chen, Hung-Pin Peng, Jhih-Wei Jian, Keng-Chang Tsai, Jeng-Yih Chang, Ei-Wen Yang, Jun-Bo Chen, Wen-Lian Hsu, An-Suei Yang*, 2012, “Protein-Protein Interaction Site Predictions with Three-Dimensional Probability Distributions of Interacting Atoms on Protein Surfaces”, PLoS One, 7(6), e37706. (SCIE)
- Chung-Ming Yu, Hung-Pin Peng, Ing-Chien Chen, Yu-Ching Lee, Jun-Bo Chen, Keng-Chang Tsai, Ching-Tai Chen, Jeng-Yih Chang, Ei-Wen Yang, Po-Chiang Hsu, Jhih-Wei Jian, Hung-Ju Hsu, Hung-Ju Chang, Wen-Lian Hsu, Kai-Fa Huang, Alex Che Ma, An-Suei Yang*, 2012, “Rationalization and Design of the Complementarity Determining Region Sequences in an Antibody-Antigen Recognition Interface”, PLoS One, 7(3), e33340. (SCIE)
- Yu-Ching Lee, Ing-Chien Chen, Chung-Ming Yu, Yi-Jen Huang, Hung-Ju Hsu, and An-Suei Yang*, 2011, “Effects of signal sequence on phage-displayed disulfide-stabilized single chain antibody variable fragment (sc-dsFv) libraries”, BIOCHEMICAL AND BIOPHYSICAL RESEARCH COMMUNICATIONS, 411(2), 348-353. (SCIE)
- Ing-Chien Chen, Chung-Ming Yu, Yu-Ching Lee, Yi-Jen Huang, Hung-Ju Hsu, and An-Suei Yang*, 2010, “Signal sequence as a determinant in expressing disulfide-stabilized single chain antibody variable fragments (sc-dsFv) against human VEGF”, MOLECULAR BIOSYSTEMS, 6(7), 1307-1315.
- Yi-Jen Huang, Ing-Chien Chen, Chung-Ming Yu, Yu-Ching Lee, Hung-Ju Hsu, Anna Ching-Ching Tung, Hung-Ju Chang, An-Suei Yang*, 2010, “Engineering Anti-vascular Endothelial Growth Factor Single Chain Disulfide-stabilized Antibody Variable Fragments (sc-dsFv) with Phage-displayed sc-dsFv Libraries”, Journal of Biological Chemistry, 285(11), 7880-7891. (SCIE)
- Hung-Ju Chang, Hung-Ju Hsu, Chi-Fon Chang, Hung-Pin Peng, Yi-Kun Sun, Hui-Ming Yu, Hsi-Chang Shih, Chun-Ying Song, Yi-Ting Lin, Chu-Chun Chen, Chia-Hung Wang, An-Suei Yang*., 2009, “Molecular Evolution of Cystine-Stabilized Miniproteins as Stable Proteinaceous Binders”, STRUCTURE, 17, 620-631. (SCIE)
- Hung-Ju Hsu, Keng-Chang Tsai, Yi-Kun Sun, Hung-Ju Chang, Yi-Jen Huang, Hui-Ming Yu, Chun-Hung Lin, Shi-Shan Mao and An-Suei Yang, 2008, “Factor-XaActive Site Substrate Specificity with Substrate Phage Display and Computational Molecular Modeling”, J. Biol. Chem, 283, 12343-12353. (SCIE)
- Chen CT, Yang EW, Hsu HJ, Sun YK, Hsu WL, Yang AS, 2008, “Protease substrate site predictors derived from machine learning on multilevel substrate phage display data.”, BIOINFORMATICS, 24(23), 2691-7. (SCIE)
- H. P. Peng and A. S. Yang, 2007, “Modeling protein loops with knowledge-based prediction of sequence-structure alignment”, Bioinformatics, 23(21), 2836-2842. (SCIE)
- Y. M. Shao, W. B. Yang, H. P. Peng, M. F. Hsu, K. C. Tsai, T. H. Kuo, A. H. Wang, P. H. Liang, C. H. Lin, A. S. Yang and C. H. Wong, 2007, “Structure-Based Design and Synthesis of Highly Potent SARS-CoV 3CL Protease Inhibitors”, ChemBioChem, 8, 1654-1657. (SCIE)
- H. Hsu, H. Cheng, H-P. Peng, S-S. Huang, M-Y. Lin and A. S. Yang, 2006, “Assessing Computational Amino Acid beta-turn Propensities with Phage-displayed Combinatorial Library and Directed Evolution”, Structure, 14, 1499-1510. (SCIE)
- R. Kuang, C. S. Leslie and A. S. Yang, 2004, “Protein backbone angle prediction with machine learning approaches”, Bioinformatics, 20, 1612-1621. (SCIE)
- A. S. Yang, L. Y. Wang, 2003, “Local structure prediction with local structure-based sequence profiles”, Bioinformatics, 19, 1267-1274. (SCIE)
- A. S. Yang, L. Y. Wang, 2002, “Local structure-based sequence profile database for local and global protein structure predictions”, Bioinformatics, 18, 1650-1657. (SCIE)
- A. S. Yang, 2002, “Structure-dependent sequence alignment for remotely related proteins”, Bioinformatics, 18, 1658-1665. (SCIE)
- A. S. Yang, B. Honig, 2000, “An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance”, J Mol Biol, 301, 665-678. (SCIE)
- A. S. Yang, B. Honig, 2000, “An integrated approach to the analysis and modeling of protein sequences and structures. II. On the relationship between sequence and structural similarity for proteins that are not obviously related in sequence”, J Mol Biol, 301, 679-689. (SCIE)
- A. S. Yang, B. Honig, 2000, “An integrated approach to the analysis and modeling of protein sequences and structures. III. A comparative study of sequence conservation in protein structural families using multiple structural alignments”, J Mol Biol, 201, 691-711. (SCIE)
- A. S. Yang, B. Honig, 1999, “Sequence to structure alignment in comparative modeling using PrISM”, Proteins, Suppl 3, 66-72. (SCIE)
- V. K. Misra, J. L. Hecht, A. S. Yang and B. Honig, 1998, “Electrostatic Contributions to the Binding Free Energy of the cI Repressor to DNA”, Biophys J, 75, 2262-2273. (SCIE)
- A. S. Yang, B. Hitz, B. Honig, 1996, “Free energy determinants of secondary structure formation: III. beta-turns and their role in protein folding”, J Mol Biol, 259, 873-882. (SCIE)
- A. S. Yang, A. S. Brill, 1996, “Thermal access to amplified chemical potential and the determination of equilibrium constants in protein solutions at subfreezing temperatures”, Biophys Chem, 58, 341-354. (SCIE)
- A. S. Yang, B. Honig, 1995, “Free energy determinants of secondary structure formation: I. alpha-Helices”, J Mol Biol, 252, 351-365. (SCIE)
- A. S. Yang, B. Honig, 1995, “Free energy determinants of secondary structure formation: II. Antiparallel beta-sheets”, J Mol Biol, 252, 366-376. (SCIE)
- B. Honig, A. S. Yang, 1995, “The free energy balance in protein folding”, Advances in Protein Chemistry, 46, 27-58.
- A. S. Yang, B. Honig, 1994, “Structural origins of pH and ionic strength effects on protein stability. Acid denaturation of sperm whale apomyoglobin”, J Mol Biol, 237, 602-614. (SCIE)
- A. S. Yang, M. R. Gunner, R. Sampogna, K. Sharp and B. Honig, 1993, “On the calculation of pKas in proteins”, Proteins, 15, 252-265. (SCIE)
- A. S. Yang, B. Honig, 1993, “On the pH Dependence of Protein Stability”, J Mol Biol, 231, 459-474. (SCIE)
- A. S. Yang, K. A. Sharp and B. Honig, 1992, “Analysis of the heat capacity dependence of protein folding”, J Mol Biol, 227, 889-900. (SCIE)
- A. S. Yang, B. Honig, 1992, “Electrostatic effects on protein stability”, Curr Opin Struct Biol, 2, 40-45.
- A. S. Yang, A. S. Brill, 1991, “Influence of the freezing process upon fluoride binding to hemeproteins”, Biophys J, 59, 1050-1063. (SCIE)
- A. S. Yang, B. J. Gaffney, 1990, “EPR Detection of Kinetic Responses to Photochemically Generated Protein Cofactors”, J Magn Reson, 90, 580-583. (SCIE)
- A. S. Yang, B. J. Gaffney, 1987, “Determination of relative spin concentration in some high-spin ferric proteins using E/D-distribution in electron paramagnetic resonance simulations”, Biophys J, 51, 55-67. (SCIE)