Original Content Source : Academia Sinica Press Releases
Autism spectrum disorder (ASD) is a pervasive neurodevelopmental and heritable complex disorder characterized by limited social communication, restricted and ritualized interests, and repetitive behavior [Mendelian Inheritance in Man (MIM) 209850]. In the US, an estimation of prevalence of ASDs was about 1 in 68 (14.6 per 1,000) children aged 8 years, suggesting that there are more than 2 million ASD individuals in the US. In Taiwan, ASD afflicts more than 15,400 people (the 2019’s report from Ministry of Health and Welfare, Taiwan); and its prevalence is increasing. Hundreds of genes affected by a variety of genomic variants have been reported to be associated with the etiology of ASD. Previous studies have provided valuable biological insights into the disorder. However, the contribution of these genetic factors to this complex disease is highly heterogeneous, with each factor accounting for a very low percentage of the general ASD population. In addition to genetic factors, environment and gene–environment interactions have been widely demonstrated to be associated with the development of ASD. Recently, using samples from post-mortem ASD and control brains, several studies have reported a wide range of dysregulation of gene expression in ASD, but the underlying molecular mechanisms remain poorly understood.
 |
| Cover legend: This image shows an autistic child who stays alone on a separate planet. This idea is derived from a documentary “Children From The Distant Planet” (2011, Taiwan), which is a famous Taiwan documentary about children with autism. Here asteroids (or musical notes) indicate microRNAs. Planetary rings imply circular RNAs, which “abnormally” attract asteroids and interfere with the so-called “normal” movement of the earth's orbit. The earth's orbit (or staff) implies mRNAs. Illustrator: Wei-Chun Hsu |
Circular RNAs (circRNAs) are a class of long non-coding RNAs produced by pre-mRNA back-splicing, which generates a structure of covalently closed loop of single-strand, non-polyadenylated circular molecules. In general, circRNAs are more stable than their corresponding co-linear mRNA isoforms. They are often expressed in temporal- and spatial-specific manners and are especially abundant in neural tissues. Several studies also showed the important role of circRNAs in development of nervous system. With the property of stability, circRNAs are likely to be useful molecules as therapeutic targets for neurological disorders. The best understood function of circRNAs is the regulatory role of microRNA (miRNA) sponges. CircRNAs thereby regulate downstream genes through mediating miRNAs. Currently, the role of circRNA-miRNA-mRNA regulatory axes in ASD remains largely unknown.
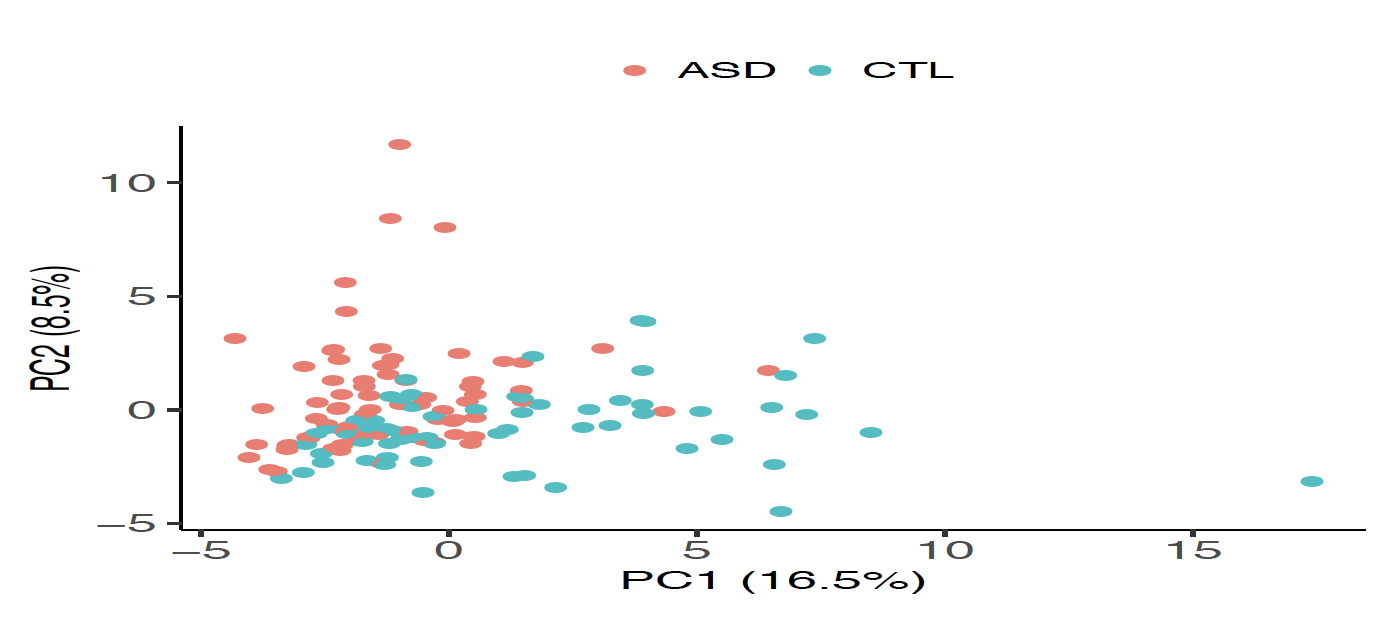 |
| Figure 1. Identification of DE-circRNAs in the ASD cortex samples. (A) The identified upregulated (22, red solid dots) and downregulated (38, green solid dots) circRNAs in the ASD cortex samples. (B) PCA based on the 60 DE-circRNAs, representing that ASD and non-ASD samples were grouped into separate clusters. |
In this study, we utilized small RNA-seq data and RNA-seq data, both of which were downloaded from the Synapse database, to investigate genome-wide circRNA expression profiling in post-mortem brains from ASD patients and non-ASD controls. We used NCLscan, which was reported to exhibit the greatest precision among currently available circRNA-detection tools, to identify circRNAs in the examined samples. We showed that circRNA expression profiles of the identified circRNAs were very similar between two types of cortex regions but were quite distinct from that of cerebellum. This result is consistent with the patterns previously observed for mRNAs and miRNAs between these brain regions. We then performed a linear mixed effects (LME) model to detect differentially expressed (DE)-circRNAs with controlling for potential confounding factors such as sex, age, brain region (FC or TC), RNA quality (RNA integrity number; RIN), host gene expression, and so on. We thus identified 60 DE-circRNAs, in which 22 were upregulated and 38 were downregulated in ASD cortex (Fig. 1A). Principal component analysis (PCA) based on the 60 DE-circRNAs revealed that ASD and non-ASD samples could be grouped into separate clusters (Fig. 1B). We also performed the weighted gene co-expression network analysis (WGCNA) to assign individual circRNAs to co-expression modules. We found that three modules were significantly correlated with ASD status. We proceeded to identify potentially ASD-associated circRNA-miRNA-mRNA regulatory axes (Fig. 2). A total of 8,170 ASD-associated circRNA-miRNA-mRNA axes were detected, which involved 2,302 target genes. We found that the target genes showed significant enrichment for autism risk genes, but not for genes implicated in monogenetic forms of schizophrenia, Alzheimer’s disease, intellectual disability, and other brain disorders. These results suggest that targets of the identified circRNA-miRNA-mRNA axes are enriched for genes causally connected with autism, but less so far genes connected with other brain disorders.
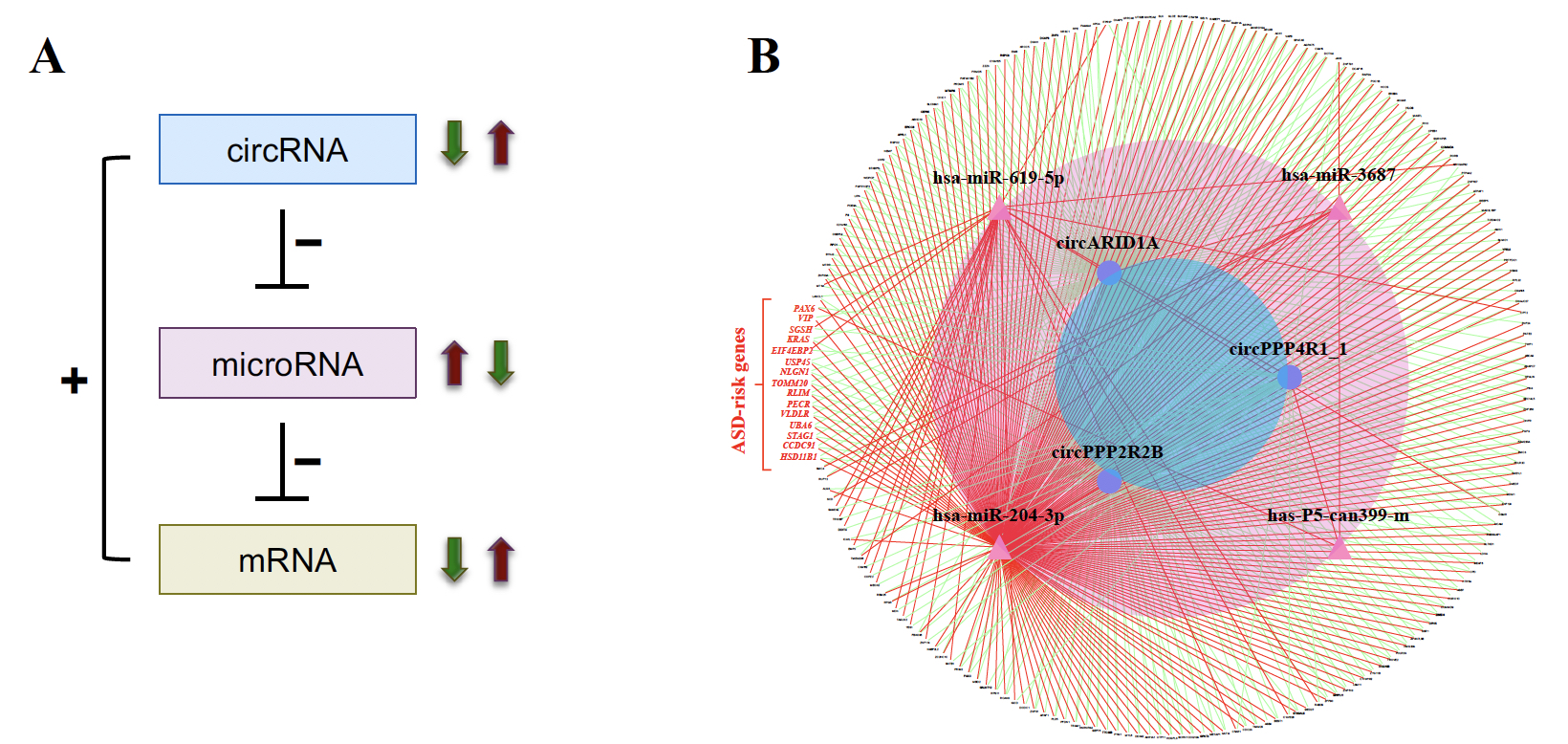 |
| Figure 2. Identification of ASD-associated circRNA-miRNA-mRNA regulatory interactions. (A) Schematic diagram representing the ASD-associated circRNA-miRNA-mRNA axes. “+” and “-” represent positive and negative correlations between the examined RNA expression, respectively. (B) The 356 upregulated circRNA-involved circRNA-miRNA-mRNA interactions. The red words represent the 12 ASD risk genes involved in the network. |
To further validate the identified circRNA-miRNA-mRNA regulatory interactions, we selected a DE-circRNA (i.e., circARID1A, which was upregulated in ASD) and confirmed that some ASD risk genes (NLGN1, STAG1, HSD11B1, VIP, and UBA6) were regulated by circARID1A via sponging a downregulated microRNA (miR-204-3p) in human neuronal cells, normal human astrocytes (NHAs) or primary human neural progenitor cells (hNPCs) (Fig. 3). We also performed circARID1A knockdown, circARID1A overexpressed, and miR-204-3p overexpression in hNPCs, respectively, and examined the fold changes of the corresponding target gene expression by microarray analysis. These results were consistent with our bioinformatics predictions.
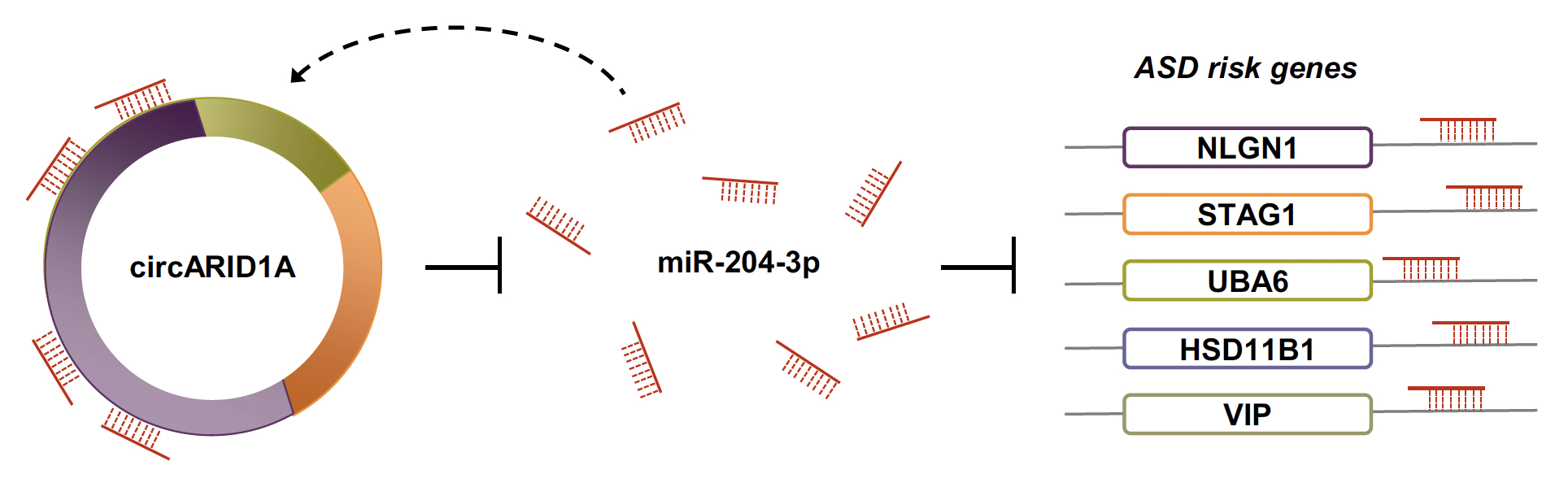 |
| Figure 3. Experimental validation of the circARID1A-regulated networks. We confirmed that circARID1A can regulated expression of five ASD risk gene (NLGN1, STAG1, HSD11B1, VIP, and UBA6) in human neuronal cells (NHAs or ReN cells) through mediating miR-204-3p. |
Taken together, our study has provided a framework for assessing the functional involvement of circRNA in ASD and the corresponding ASD-associated circRNA-miRNA-mRNA regulatory axes. By integrating ASD candidate gene sets and circRNA, miRNA, and mRNA dysregulation data derived from the same ASD cortex samples, we have provided a rich set of ASD-associated circRNA candidates and the corresponding circRNA-miRNA-mRNA axes, particularly those involving ASD risk genes, for further investigation in ASD pathophysiology. To the best of our knowledge, this is the first systems-level view of landscape of circRNA regulatory networks in ASD cortex samples. The proposed framework offers further analysis of circRNAs in other human complex diseases such as Alzheimer’s disease, Parkinson's disease, and schizophrenia.
This study integrates medical big data analysis with systems and molecular biology (Fig. 4). The co-first authors of this manuscript include Yen-Ju Chen (Ph.D. student/molecular biology), Chia-Ying Chen (postdoc/biostatistics), Te-Lun Mai (postdoc/biophysics). The corresponding author is Prof. Trees-Juen Chuang. All these authors work in the Prof. Chuang’s group in Genomics Research center, Academia Sinica. The manuscript has been published in Genome Research in March 2020 (see https://genome.cshlp.org/content/30/3/375.full.pdf+html). Other related studies of Prof. Chuang’s group include:
(1) NCLscan and the related circRNA-detection tools, which exhibit the greatest precision among currently available circRNA-detection tools (Nucleic Acids Research, 44(3), e29, 2016;PLoS Comput Biol., 15, e1006158, 2019;BMC Bioinformatics, 20:3, 2019).
(2) Investigation of circRNA and trans-spliced RNA biogenesis (Nucleic Acids Research, 46 (7): 3671 3691, 2018).
 |
| Figure 4. The radar chart representing the importance of the study on each of the six domains of knowledge. |