Compounding Informatics and Stem Cell studies, Dr. Trees-Juen Chuang of Genomics Research Center teamed up with Dr. Hung-Chih Kuo of Institute of Cellular and Organismic Biology, had successfully created a pipeline named “TSscen” to systematically identify trans-splicing in human embryonic stem cells (ESCs). TSscan comprises computational algorithms and filters, Next Generation Sequencing technologies, and laboratory bench mark tests with human ESCs. The group did not only prove the accuracy of TSscan in finding true trans-spliced RNAs, they have also proved one of the pinpointed RNA called tsRMST is a crucial element toward the pluripotency in human ESCs. Plurpotency is a stem cell's ability to differentiate into different cell types. This study has been published online in the “Genome Research” Journal in October, and was reported in the Research Highlights section of the “Nature Reviews Genetics” Journal right afterwards.
The codes carried in DNA have to go through a series of stages, include transcription, translation, post-translational modification, then, a functional protein can be made. In the course of RNA transcription, it is similar to a film editing task, where when clips of takes are put together by cut and paste. Of note, transcription is the synthesis of RNA from a DNA template, in which the intronic sequences will be removed from premature message RNAs by the so-called RNA splicing process.
As there are more and more Genome studies, scientists are noticing the existence of trans-spliced RNAs. These RNAs are called trans-splicing, because it is observed that their sequences somewhat come from two or more separate precursor mRNAs. Some of the trans-spliced RNAs may have sequences that are orderly inconsistent with their corresponding DNA template, some may even have sequences from different genes. Recently, the next-generation sequencing (NGS) techniques, which have brought an unprecedented opportunity for globally investigating transcriptomes, enable many bioinformatics methods to be developed and detect thousands of non-co-linear transcript candidates in varied species. Trans-splicing events detected by such means may, however, include a considerable number of false positives that arise from genetic rearrangements and experimental artifacts, template switching especially. Therefore, only a few trans-spliced transcripts have been well-verified or well-documented in higher eukaryotes to date.
To address this, the research team developed a new pipeline, TSscan, which integrated different types of high-throughput long-/short-read transcriptome sequencing of different human ESC lines to effectively minimize false positives while detecting trans-splicing. Upon completion of the TSscan screening process, almost 99.9% of ~9,000 candidates were eliminated and 9 possible trans-spliced RNAs were finally identified. Of note, further analyses revealed that the majority of the filtered candidates were artifacts. In the following work, the research team conducted the validation tasks by using human ESCs. And, in the end, final validated trans-spliced RNA candidates were narrowed down to only 4.
On top of that, by investigating multipe human ESC lines, the research team showed that these four trans-spliced RNAs were all highly expressed in in human pluripotent stem cells including ESCs and iPSCs (induced Pluripotent Stem cells), and differentially expressed during human ESC differentiation. These results suggested that these identified trans-spliced RNAs could have roles in human ESC pluripotency and early human embryonic development.
Most interesting is that, one of the four trans-spliced RNAs named tsRMST, which could be the first reported trans-spliced large intergenic noncoding RNA so far, was proved to be specifically expressed in pluripotent cells. The team disrupted tsRMST expression in ESCs, and observed that without tsRMST, the cell differentiation starts. Therefore, tsRMSTcould be a key factor that suppresses the differentiation and maintain the pluripotency of human ESCs (Fig. 1).
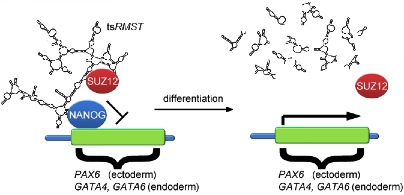
Figure 1. A putative model for regulation of gene expression by tsRMST in pluripotent stem cells. PAX6、GATA4、and GATA6 are key lineage-specific transcription factors for reorganization of ESC lineage differentiation.
Take together, this study not only describes a new approach to systematically detect trans-splicing, but also provides further insight into the potential roles of trans-splicing in human ESC pluripotency and early human embryonic development.
Postdoctoral fellows Dr. Chan-Shu Wu and Dr. Chun-Ying Yu are the co-first authors for bioinformatics and stem cell studies, respectively. This study was sponsored by Academia Sinica and NSC.
Reference sites:
Genome Research: http://genome.cshlp.org/content/early/2013/10/16/gr.159483.113.long
Nature Reviews Genetics: http://www.nature.com/nrg/journal/vaop/ncurrent/full/nrg3618.html